Protein purification
Prior to each protein analysis the target protein has to be separated from a complex mixture to focus on the function without influences from other molecules, making protein purification to one of the main applications in biotechnology. A target protein can be separated from other molecules via specific properties like molecular mass, charge, hydrophobicity or affinity to another molecule, whereas the latter the most efficient property is. The molecular mass, charge and hydrophobicity can be similar between different proteins, but the affinity to another molecule is a unique property.
To purify a protein with the help of their affinity to another molecule, the interacting partner (ligand), e.g., a protein, a small molecule, or a metal, is immobilized on the stationary phase of the chromatography matrix. The stationary phase mostly consists of agarose or synthetic polymers and is packed into a column in the form of beads. The beads are surrounded and moisturized by a liquid, called the mobile phase. When the target protein-containing sample is applied, it enters the mobile phase and runs through the beads of the stationary phase. Meanwhile, the target protein can bind to the ligand, whereas other molecules remain in the mobile phase and can be removed by washing. For elution of the target protein, the interaction to the ligand is resolved by changing the buffer conditions, for example, the pH value, or by adding a specific competitor that supplants the target protein from the ligand resulting in dissociation of the target, while the ligand remains immobilized on the stationary phase.
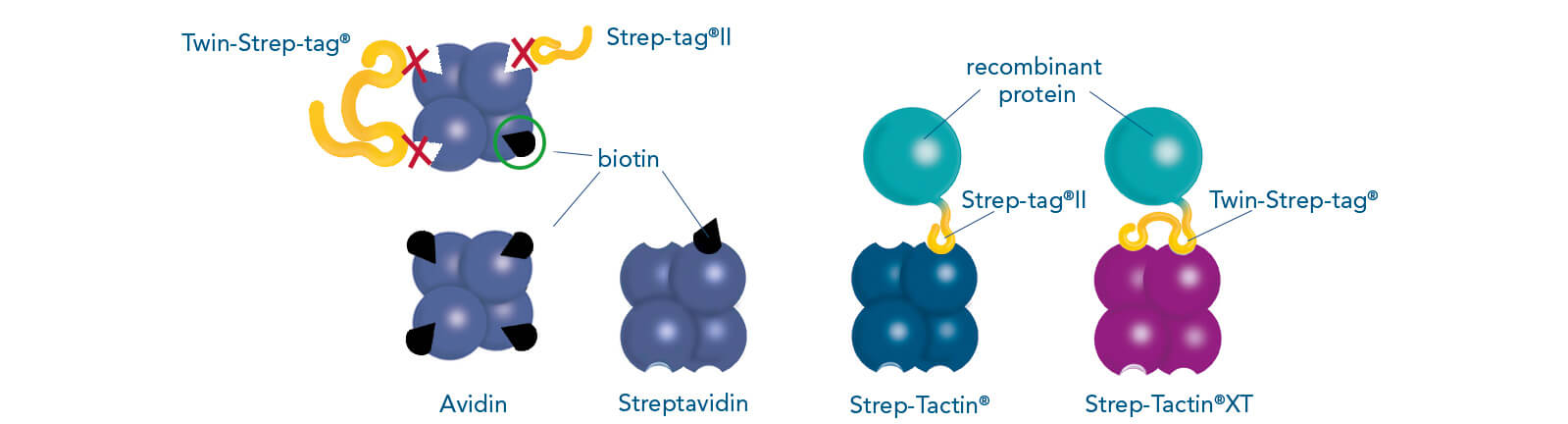
Such a direct affinity purification strategy is most commonly used for antibodies based on antigen-antibody interactions. One example is protein A (ligand), which can be utilized to purify immunoglobulin G antibodies (target protein). But how can a protein be purified if an interaction partner is not known so far? In this case, the affinity of known interactions can be utilized for the indirect capture of the target protein. For this purpose, the target protein is tagged with a short peptide of one of the known interactors, which can bind to the other interaction partner immobilized on the stationary phase. The short peptide of the known interactor is called affinity tag and can be, for instance, the His-tag, GST-tag, Strep-tag®II, or Twin-Strep-tag®. They can bind to ligands such as metal ions, glutathione, Strep-Tactin®, or Strep-Tactin®XT, respectively.
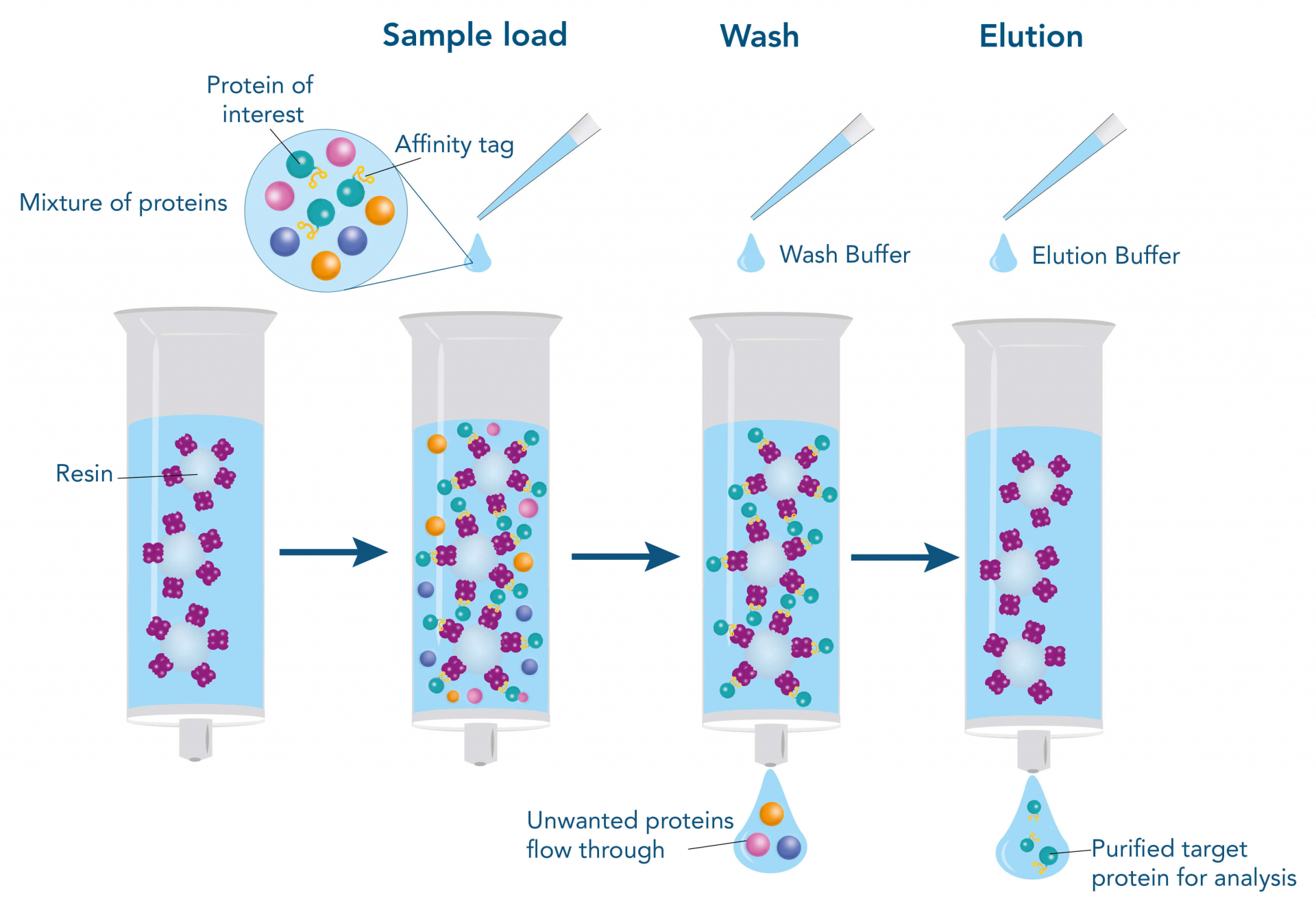

Due to the highly specific selection and the resulting purity of target proteins, affinity-based systems have become the flagship method of purification. However, some of the widely used affinity tags, such as the His-tag, exhibit several drawbacks and limitations, since they can increase the risk of distorting the natural conformation of the target protein or necessitate stringent elution and wash conditions affecting the yield of the target protein. Additionally, many tags are not compatible with varying buffer conditions and must be removed to not impair downstream processing.
The widely used Strep-tag® technology, consisting of two engineered streptavidin variants, Strep-Tactin® and Strep-Tactin®XT, and two affinity tags, Strep-tag®II and Twin-Strep-tag®, is not limited in the use of buffers and its high specificity leads to obtaining highly pure proteins.
Therefore, IBA provides several resins, either coupled with Strep-Tactin® or Strep-Tactin®XT, which are all applicable for Strep-tag®II and Twin-Strep-tag® fusion protein purification. The purification cycle varies between both streptavidin variants, but both resin types serve the same goal: simple, fast, and variable protein purification procedures for highly pure proteins.
The special differences to other systems, especially between the His-tag system and Strep-tag® technology, are presented in a comprehensive comparison. It summarizes the differences as well as recommends one of the systems depending on the properties of the target protein, expression host, and purification conditions.
Antibody purification
Antibodies are versatile tools in biotechnology. They can be utilized for the direct and indirect detection of antigens in various assays, as capture molecule immobilized on microplates, or as ligand for protein purification. However, before they can be deployed, they have to be purified from cell lysates, cell culture supernatants, or biological probes. The purification can take place without affinity tag via the physicochemical properties, antigen-specific affinity or the antibody class. Physicochemical properties are the molecular weight, charge, or clusters of specific residues. Depending on the origin of the sample, purification with the help of physicochemical properties or antigen-specific affinity can lead to the isolation of further proteins and antibodies with similar properties besides the target antibody. Thus, if a specific antibody with a high purity should be obtained, antibody class-specific affinity chromatography is recommended.
One of the ligands used for antibody class-specific affinity chromatography is Protein A. Originally, Protein A is a surface protein of the Staphylococcus aureus cell wall, which can bind immunoglobulins (antibodies) within the Fc region of their heavy chain without regard to antigen specificity. It is composed of five homologous Ig-binding domains that fold into a three-helix bundle. The IgG binding ability was utilized to produce an affinity tag-free system for direct purification of IgGs from serum, cell supernatants as well as extracts. The Protein A affinity chromatography resin captures IgGs from various mammalian species with different affinities under physiological buffer conditions (pH 7.2–7.4), whereas other molecules and antibodies flow through. Afterwards, the IgGs are released from capture by reduction of the pH to a more acidic value (pH 2.7). IBA’s Protein A affinity chromatography resin consists of a highly cross-linked agarose coupled with recombinant Protein A expressed in E. coli and is available as cartridge for HPLC/FPLC, gravity flow columns, and 50% suspension to allow the use in various purification applications.
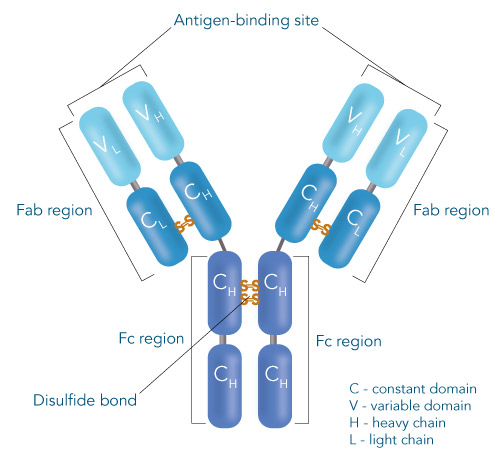

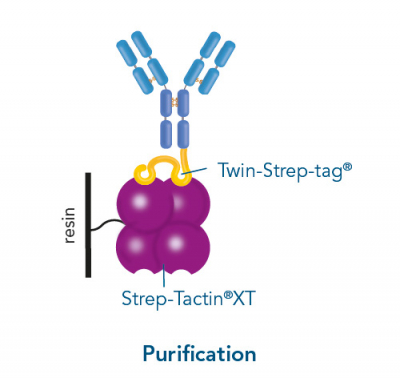
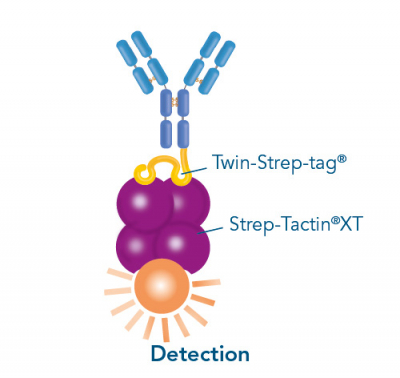
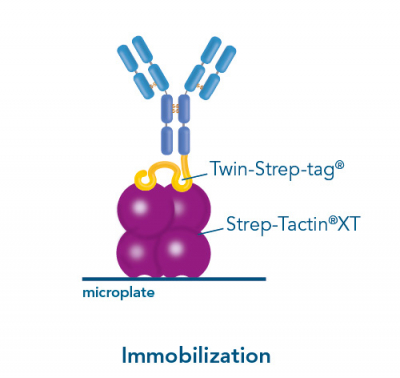
Usually, an affinity tag-free purification method is preferred. However, sometimes an affinity tag-based purification procedure can be beneficial, especially if the antibody is to be implemented in subsequent immobilization or detection. For this purpose, the application of the Strep-tag® technology is highly recommended. The antibody can be simply isolated in high purity with a resin perfectly suitable for large proteins, Strep-Tactin®XT 4Flow®. After removing biotin, the eluent, from the elution fraction via dialysis or size exclusion, the antibody can be immobilized on Strep-Tactin®XT-coated microplates or labeled with a Strep-Tactin®XT conjugate and applied for detection assays.
Biotin blocking
Blocking of free biotin in cell culture supernatants and lysates
Cell culture supernatants often contain high amounts of free biotin. These are unproblematic when working with Strep-Tactin®XT resins, since biotin does neither bind irreversibly to this ligand nor reduces the binding capacity. However, this is the case for the application of resins coupled to Strep-Tactin®. To prevent the impairment of Strep-Tactin®, BioLock, which contains avidin, can be applied to mask free biotin. Avidin is a homolog of streptavidin and can be found in the egg white of birds, reptiles, and amphibians. It consists of four subunits, each of which can bind one molecule of biotin. In contrast to the streptavidin variants, Strep-Tactin® and Strep-Tactin®XT, it is not able to bind Strep-tag®II or Twin-Strep-tag® fusion proteins. Therefore, avidin can be used to selectively mask free biotin in cell lysates or cell culture supernatants, whereas the Strep-tag®II or Twin-Strep-tag® at the target protein remain accessible. An overview of media containing biotin and cell-internal biotin pools is given in the manual for biotin blocking. Blocking of biotin by BioLock is simple and fast. Only a small amount has to be added and, after a short incubation, the sample is ready for protein purification.
Blocking of biotinylated proteins
Besides free biotin, cell lysates also contain small amounts of biotinylated proteins. Normally, these proteins do not influence the purification results with Strep-Tactin® or Strep-Tactin®XT due to their low abundance. However, when it comes to analytic applications with high sensitivity, impurities should be prevented. One option is the treatment of the sample with BioLock prior to protein purification. Otherwise, if the target protein is already purified and should be detected via western blot with Strep-Tactin® or Strep-Tactin®XT conjugates, the application of Biotin Blocking Buffer is possible. Just as BioLock, Biotin Blocking Buffer contains avidin to mask biotin and biotinylated proteins, but the buffer is especially optimized for western blots. Shortly before detection, the membrane is incubated with the diluted buffer for a few minutes. However, if the detection occurs with the aid of StrepMAB-Classic or StrepMAB-Immo, masking of biotinylated proteins is not necessary. In contrast to Strep-Tactin® or Strep-Tactin®XT conjugates, these antibodies do not bind to biotin or biotinylated proteins and therefore do not lead to unspecific signals.